Python script
How to create a Python script for multiple datasets execution
This code is an implementation of a Python main script. It demonstrates how to read data from files, process it, and generate results in different formats.
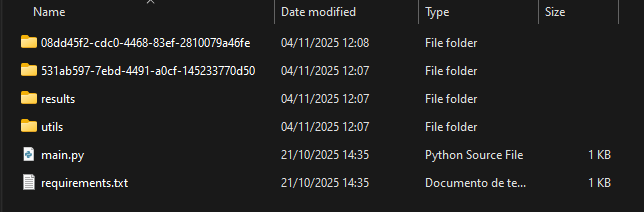
In this example, we will have this directory structure:
|-- utils/| |-- cluster.py|-- main.py|-- requirements.txt- The
main.pyandrequirements.txtfiles are mandatory and must have these specific filenames. - Other files can be added to use different Python modules; in this example, we are using the
utilsdirectory. These modules can have any name and directory structure, as long as they are correctly loaded in themain.pyfile. - The
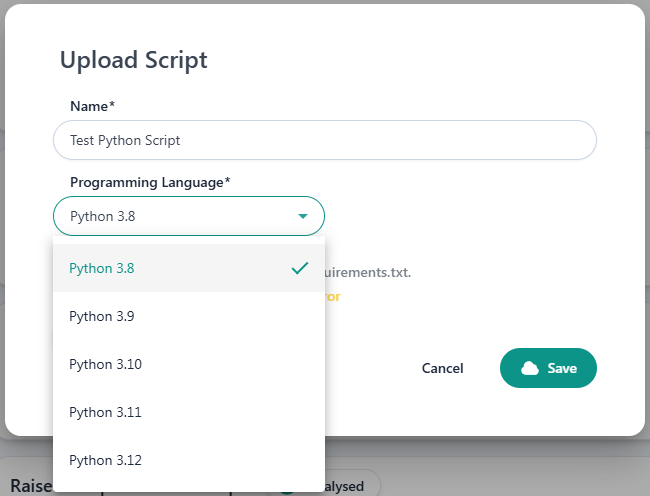
requirements.txtfile should list all the required library versions for execution. All of them must be compatible with the Python runtime version selected for the experiment:3.8,3.9,3.10,3.11or3.12.
Import Required Libraries
There are no specific required libraries, you can import any library for your own use: e.g. pandas. And, also import other Python modules from your files.
import jsonfrom dotenv import load_dotenvimport pandas as pdfrom utils.cluster import cluster_data,plot_clusters_figureImport Data Files (shareable samples)
Only when working locally, you’ll have to download and organize your data files according to the required folder structure. (see also Dynamic Dataset(s) Accessing.)
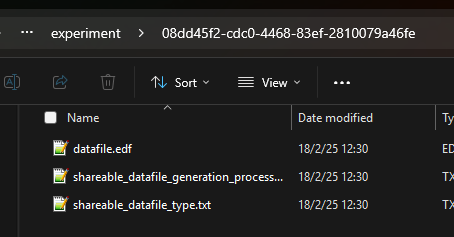
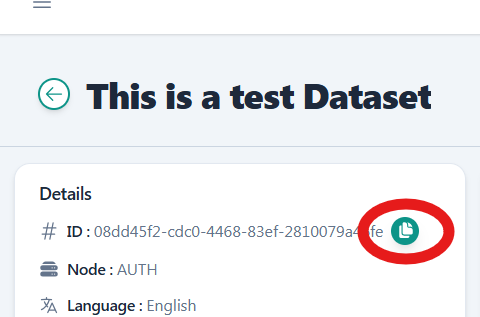
Each data file should be stored in a folder named after its ID (the data file’s UUID can be found in its metadata) in the same directory with the main.py script.
You can copy the dataset ID from the dataset details page on the RAISE portal.
Then, create a .env file that defines the variable RAISE_DATASET_ID_LIST. This variable should contain an ordered list of the dataset UUIDs in the following format:
RAISE_DATASET_ID_LIST=["00000000-0000-0000-0000-000000000001","00000000-0000-0000-0000-000000000002"]Read Data Files
Once your experimental environment is ready, you can read the files:
load_dotenv()
dataset_ids = json.loads(os.getenv("RAISE_DATASET_ID_LIST"))
dataset_id_0 = dataset_ids[0]dataset_id_1 = dataset_ids[1]
datafile_extension_0 = "csv"datafile_extension_1 = "csv"
file_path_0 = f"{dataset_id_0}/datafile.{datafile_extension_0}"file_path_1 = f"{dataset_id_1}/datafile.{datafile_extension_1}"The data file must always be named “datafile” with the appropriate extension: e.g., .csv, .txt, .edf, .json…
There is no limit to the number of datasets that can be loaded.
Use different functions to load data depending on the data type. For example, if you have CSV files, you can use the read_csv from pandas:
data_csv_0 = pd.read_csv(file_path_0)data_csv_1 = pd.read_csv(file_path_1)
data = pd.concat([data_csv_0, data_csv_1], ignore_index=True).sort_values(by=data_csv_0.columns[0])Run the Code
In this section, you can add the code to process the data. The code will depend on the specific task you want to perform (e.g., clustering, analysis, etc.). Here’s an example:
clustered_data, reduced_clustered_data, cluster_centers = cluster_data(data=data)clusters_figure = plot_clusters_figure(data=reduced_clustered_data, centroids=cluster_centers)Gather Results
Any type of file can be taken as a result -> images, CSV, text… Moreover, the number of results is not limited. It is worth noting that the results must now be stored under the “results” directory (it is at the same level as the main file execution).
os.makedirs("results", exist_ok=True)clustered_data.to_csv("results/clustered_data.csv")reduced_clustered_data.to_csv("results/reduced_clustered_data.csv")Complete main.py script (download)
# Import Required Librariesimport jsonfrom dotenv import load_dotenvimport pandas as pdfrom utils.cluster import cluster_data,plot_clusters_figure
# NOTE: Remember to prepare the environment for local execution (see the "Import Data Files" section above)
# Read Data Filesload_dotenv()
dataset_ids = json.loads(os.getenv("RAISE_DATASET_ID_LIST"))
dataset_id_0 = dataset_ids[0]dataset_id_1 = dataset_ids[1]
datafile_extension_0 = "csv"datafile_extension_1 = "csv"
file_path_0 = f"{dataset_id_0}/datafile.{datafile_extension_0}"file_path_1 = f"{dataset_id_1}/datafile.{datafile_extension_1}"
data_csv_0 = pd.read_csv(file_path_0)data_csv_1 = pd.read_csv(file_path_1)
data = pd.concat([data_csv_0, data_csv_1], ignore_index=True).sort_values(by=data_csv_0.columns[0])
# Run the Code# Add here the code to run. You can create different functions and the results should be clear stated.clustered_data, reduced_clustered_data, cluster_centers = cluster_data(data=data)clusters_figure = plot_clusters_figure(data=reduced_clustered_data, centroids=cluster_centers)
# Gather Resultsos.makedirs("results", exist_ok=True)clustered_data.to_csv("results/clustered_data.csv")reduced_clustered_data.to_csv("results/reduced_clustered_data.csv")requirements.txt file
dotenvLogs
Finally, the log system has been improved. In the event that the experiment does not fail, the user’s “prints()” in the script will be logged. In the case of an error, the logs save the exact error that caused the execution to fail.
In case the main.py execution fails, you will be able to see the exact reason of the failure (wrongly defined variables, unexpected indentations…).
In the case where the creation of the child container is not successful the logs will contain the reason for the failure (incompatible versions in the requirements txt, non-existing package versions…).\
You can find some examples at the templates section.